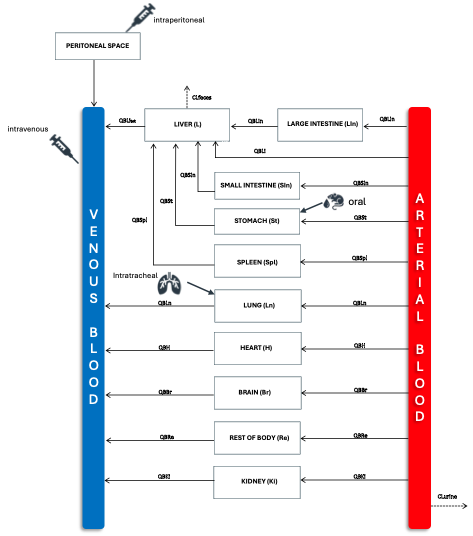
The model applied here is an implementation of a perfusion-limited physiologically based kinetic (PBK) model describing the biodistribution of Graphene Oxide (GO) in mice. It was developed as part of the 2D materials CHIASMA demo-case, building on experimental biodistribution data and physiological parameters available in the literature and PK-Sim database.
Graphene Oxide is a two-dimensional nanomaterial composed of layered carbon sheets arranged in a honeycomb lattice, enriched with oxygen-containing functional groups such as hydroxyl (-OH) and carboxyl (-COOH) (Magne et al., 2021). These features confer exceptional physicochemical properties that have positioned GO as a promising material for biomedical and energy applications. However, its biodistribution and systemic fate are highly dependent on its physicochemical characteristics—size, surface chemistry, agglomeration state, and functionalization—as well as the route of exposure (oral, inhalation, dermal, or parenteral). Consequently, the model was designed to capture how these determinants influence GO disposition across biological systems.
The model was constructed on published biodistribution data (i.e. six different biodistribution studies), incorporating organ compartments for which GO mass or concentration values were reported. Specifically the extracted data included organ-specific single time-point or time-course profiles in blood, kidney, liver, stomach, small and large intestine, lung, heart, spleen, and brain, following single intravenous, oral, intratracheal, or intraperitoneal administration. The list of the biodistribution studies used for models' calibration are presented in Table 1. All compartments are interconnected via arterial and venous blood flows, representing the systemic circulation. The model structure and flow scheme are shown below.
The model equations describe the uptake, distribution, and clearance of GO under various exposure conditions (intravenous, oral, intratracheal, and intraperitoneal). GO uptake in each organ compartment is governed by tissue:blood partition coefficients and perfusion-limited kinetics. As an initial step, a deterministic parameter estimation was conducted to derive organ-specific partition coefficients, absorption, and clearance rates by simultaneously fitting the model to multiple experimental datasets.In the first phase, we used one common set of chemical-specific parameters for all GO types and exposure routes to ensure mechanistic consistency. This generic PBK model generated organ and blood time-course profiles by converting predicted GO amounts into concentrations using physiological organ volumes, enabling straightforward comparison with published biodistribution data.
In the second phase, we fitted the model individually to each biodistribution study dataset. This allowed us to estimate study-specific partition coefficients (PC), absorption and clearance rates, better reflecting the differences in GO formulations, such as size, coating, and administration route. Study-specific models resulted in considerably improved fits when GO physicochemical properties and exposure conditions were implicitly considered.
The R implementation of the model is available through the Jaqpot platform and in an open-access GitHub repository. This repository hosts multiple published PBK models for various substances and organisms, supporting transparent reuse and further development within the nano-PBK modelling community.
Table 1. Graphene oxide biodistribution studies in mice
| Study No | Reference | GO Material | Route | Dose (mg/kg BW) | Tissues | Sex |
|---|---|---|---|---|---|---|
| 1 | Yang et al. (2010) | Nanosheets | iv | 4 | blood, liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 2a | Liu et al. (2012) | Small sheets | iv | 1, 2, 10 | blood, liver, spleen, kidneys, heart, lungs, stomach, small intestine, large intestine, brain | M |
| 2b | Liu et al. (2012) | Large sheets | iv | 1, 2, 10 | blood, liver, spleen, kidneys, heart, lungs, stomach, small intestine, large intestine, brain | M |
| 3a | Yang et al. (2013) | PEGylated nanosheets | ip | 50 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 3b | Yang et al. (2013) | PEGylated nanosheets | oral | 100 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 3c | Yang et al. (2013) | PEGylated reduced sheets | ip | 50 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 3d | Yang et al. (2013) | PEGylated reduced sheets | oral | 100 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 3e | Yang et al. (2013) | PEGylated nanoreduced sheets | ip | 50 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 3f | Yang et al. (2013) | PEGylated nanoreduced sheets | oral | 100 | liver, spleen, kidneys, heart, lungs, stomach, intestine, brain | F |
| 4 | Li et al. (2013) | Nanosheets | in | 10 | blood, liver, spleen, kidneys, heart, lungs, stomach, small intestine, large intestine | M |
| 5a | Li et al. (2014) | Nanosheets | iv | 5 | blood, liver, spleen, kidneys, heart, lungs, stomach, small intestine, large intestine | M |
| 5b | Li et al. (2014) | PEGylated nanosheets | iv | 5 | blood, liver, spleen, kidneys, heart, lungs, stomach, small intestine, large intestine | M |
| 6a | Mao et al. (2016) | Few-layer GO particles | in | 5 | liver, spleen, lungs, stomach, small intestine, large intestine, feces | M |
| 6b | Mao et al. (2016) | Few-layer GO particles | oral | 10 | liver, spleen, lungs, stomach, small intestine, large intestine, feces | M |
Abbreviations:
iv = intravenous · ip = intraperitoneal · in = intranasal · M = male · F = female